PSICHIC: physicochemical graph neural network for learning protein-ligand interaction fingerprints from sequence data
Abstract
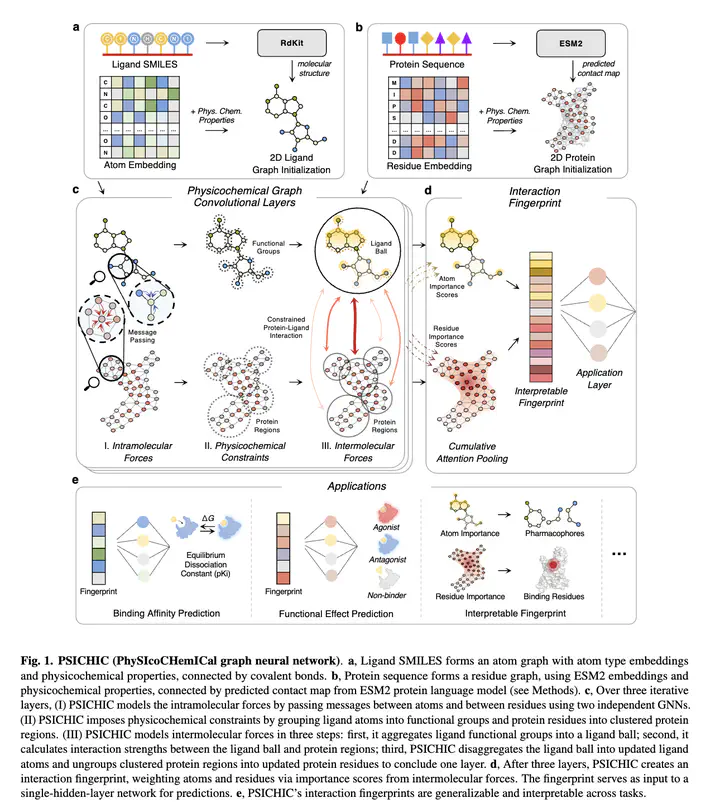
In drug discovery, determining the binding affinity and functional effects of small-molecule ligands on proteins is critical. Current computational methods can predict these protein-ligand interaction properties but often lose accuracy without high-resolution protein structures and falter in predicting functional effects. We introduce PSICHIC (PhySIcoCHemICal graph neural network), a framework uniquely incorporating physicochemical constraints to decode interaction fingerprints directly from sequence data alone. This enables PSICHIC to attain first-of-its-kind emergent capabilities in deciphering mechanisms underlying protein-ligand interactions, achieving state-of-the-art accuracy and interpretability. Trained on identical protein-ligand pairs without structural data, PSICHIC matched and even surpassed leading structure-based methods in binding affinity prediction. In a library screening for adenosine A1 receptor agonists, PSICHIC discerned functional effects effectively, ranking the sole novel agonist within the top three. PSICHIC’s interpretable fingerprints identified protein residues and ligand atoms involved in interactions. We foresee PSICHIC reshaping virtual screening and deepening our understanding of protein-ligand interactions.